\(\renewcommand\AA{\mathring{A}}\)
SliceViewer Testing¶
Introduction¶
The Sliceviewer in Workbench has the joint functionality of the SpectrumViewer and SliceViewer from MantidPlot. So while the advanced use cases for multi-dimensional diffraction data are important to test, it is also worth checking more basic uses, for example opening a Workspace2D and examining the subplots and dynamic cursor data.
See here for a brief overview of the Sliceviewer.
Basic Usage¶
MatrixWorkspace¶
Do the following tests with an EventWorkspace (e.g. CNCS_7860_event.nxs) and a Workspace2D (e.g. MAR11060.raw) from the TrainingCourseData.
Load the workspace and open in sliceviewer (right-click on the workspace in the ADS > Show Slice Viewer).
Confirm that for MatrixWorkspaces the Peak overlay, Nonorthogonal view and Non-axis aligned cutting tool buttons should be disabled (greyed out).
Check the other toolbar buttons.
Pan and Zoom (use mouse scroll and magnifying glass tool) and Home
As you zoom check the color scale changes if autoscale checked in checkbox below colorbar).
Toggle grid lines on/off
Enable line plots
Confirm the curves update with the cursor position correctly
Export cuts to workspaces in ADS using keys x, y, c
This should produce workspaces in the ADS with suffix _cut_x and _cut_y
Plot these in workbench check they agree with sliceviewer plots
Disable line plots and Enable ROI tool
Draw, move and resize the rectangle
Move it off the axes (it should just clip itself to be contained within the axes).
Export the cuts with keys x, y, c
In addition the ROI can be exported by pressing r
This should produce another workspace with suffix _roi
Open it in sliceviewer and check the data and limits agree with the ROI drawn.
Disable the ROI tool
The line plot tool should remain enabled.
Try saving the figure (with and without ROI/lineplots).
Test the colorbar and colorscale
Change normalisation
The color limits should only change if autoscale is enabled.
Change the scale type to e.g. Log
In Log scale bins with 0 counts should appear white
When you zoom in to a region comprising only of bins with 0 counts it will set the color axis limits to (0.1,1) and force the scale to be linear
Change colormap
Reverse colormap
Change auto-scale option from default Min/Max to another e.g. 3-sigma
Test transposing axes: click the Y button to the right of the Time-of-flight label (top left corner) - the image should be transposed and the axes labels updated.
Test the Cursor Information Widget (table at top of sliceviewer window with TOF, spectrum, DetID etc.)
Confirm it tracks with the cursor when Track Cursor is checked
Uncheck the track cursor and confirm it updates when the cursor is clicked.
Transpose the axes (as in step 6) and click on the colorfill plot. Confirm the clicked cursor position is correctly displayed in the table. If Track Cursor is checked, the table should still track the cursor correctly.
Resize the sliceviewer window, check the widgets, buttons etc. are still visible and clear for reasonable aspect ratios.
MD Workspaces¶
MD workspaces hold multi-dimensional data (typically 2-4D) and come in two forms: MDEventWorkspace, MDHistoWorkspace.
In terms of sliceviewer functionality, the key difference is that MDHistoWorkspace have binned the events onto a regular grid and cannot be dynamically rebinned unless the original MDWorkspace
(that holds the events) exists in the ADS (and the MDHistoWorkspace has not been altered by a binary operation e.g. MinusMD).
MDWorkspace (with events)¶
Create a 3D and 4D MDWorkspaces with some data - repeat the following tests with both
md_4Dandmd_3D. Note this will throw a warning relating to theSlicingAlgorithm.
from mantid.simpleapi import *
md_4D = CreateMDWorkspace(Dimensions=4, Extents=[0,2,-1,1,-1.5,1.5,-0.25,0.25], Names="H,K,L,E", Frames='HKL,HKL,HKL,General Frame',Units='r.l.u.,r.l.u.,r.l.u.,meV')
FakeMDEventData(InputWorkspace=md_4D, UniformParams='5e5') # 4D data
FakeMDEventData(InputWorkspace=md_4D, UniformParams='1e5,0.5,1.5,-0.5,0.5,-0.75,0.75,-0.1,0.1') # 4D data
# Add a non-orthogonal UB
expt_info = CreateSampleWorkspace()
md_4D.addExperimentInfo(expt_info)
SetUB(Workspace='md_4D', c=2, gamma=120)
# make a 3D MDEvent workspace by integrating over all E
md_3D = SliceMD(InputWorkspace='md_4D', AlignedDim0='H,0,2,100', AlignedDim1='K,-1,1,100', AlignedDim2='L,-1.5,1.5,100')
# Create a peaks workspace and fake data in 3D MD
CreatePeaksWorkspace(InstrumentWorkspace='md_3D', NumberOfPeaks=0, OutputWorkspace='peaks')
CopySample(InputWorkspace='md_3D', OutputWorkspace='peaks', CopyName=False, CopyMaterial=False, CopyEnvironment=False, CopyShape=False)
AddPeakHKL(Workspace='peaks', HKL='1,0,1')
AddPeakHKL(Workspace='peaks', HKL='1,0,0')
Test the toolbar buttons pan, zoom, line plots, ROI as in step 3 of the MatrixWorkspace instructions.
This workspace should be dynamically rebinned - i.e. the number of bins within the view limits along each axis should be preserved when zooming.
The cursor info table should also display the HKL of the cursor
Change the number of bins along one of the viewing axes (easier to pick a small number e.g. 2)
Change the integration-width/slice-thickness (spinbox to the left of the word thick) along the non-viewed axes.
Increasing the width should improve the stats on the uniform background and the color limit should increase (event counts are summed not averaged).
Change the slicepoint along one of the non-viewed axes
Confirm the slider moves when the spinbox value is updated.
Confirm moving the slider updates the spinbox.
Check the slicepoint is updated in the cursor info table (note if Track Cursor is checked you will need to click on the colorfill plot again).
Test the Nonorthogonal view

Click the nonorthogonal view button in the toolbar
This should disable ROI and lineplot buttons in the toolbar
This should automatically turn on gridlines
When H and K are the viewing axes the gridlines should not be perpendicular to each other
The features in the data should align with the grid lines
Zoom and pan
Confirm the autoscaling of the colorbar works in non-orthogonal view
Note the range of data in X and Y axes to be 0 to 2 for H and -1 to +1 for K respectively. Swap the viewing axes by clicking Y button next to H in the top left of the window
X and Y axes should now display data for K and H respectively preserving their original ranges.
Click on X button next to the H button to swap the axes again
Now X and Y axes should display data for H and K respectively preserving their original ranges.
Change one of the viewing axes to be L (e.g. click X button next to L in top left of window)
Gridlines should now appear to be orthogonal
For
md_4Donly change one of the viewing axes to be E (e.g. click Y button next to E in top left of window)Nonorthogonal view should be disabled (only enabled for momentum axes)
Line plots and ROI should be enabled
Change the viewing axis presently selected as E to be a momentum axis (e.g. H)
The nonorthogonal view should be automatically re-enabled.
Test the Peak Overlay

Click to peak overlay button in the toolbar
Check the Overlay? box next to
peaksThis should open a table (peak viewer) on the RHS of the sliceviewer window - it should have two rows corresponding to peaks at HKL = (1,0,1) and (1,0,0).
Double click a row
It should change the slicepoint along the integrated momentum axis and zoom into the peak - e.g. When double clicked on the row with HKL = (1,0,1) or HKL = (1,0,0) the slicepoint along L will be set to 1 or 0 respectively, and there will be a cross at (X,Y) = (H,K) = (1,0) in both cases.
Note for
md_4Da peak will not be plotted if a non-Q axis is viewed, if both axes are Q-dimensions then the cross should be plotted at all E (obviously a Bragg peak will only be on the elastic line but the peak object has no elastic/inelastic logic and the sliceviewer only knows that E is not a momentum axis, it could be temperature etc.).
Click Add Peaks in the Peak Actions section at the top of the peak viewer (note for 4D workspaces the peak overlay won’t work for non_Q dimensions).
Click somewhere in the colorfill plot
Confirm a peak has been added to the table at the position you clicked
Note that peaks need to have H > 0 to be valid due to the assumed beam direction (otherwise you will get an error in the log
ValueError: Peak::setQLabFrame(): Wavelength found was negative)
Click Remove Peaks
Click on the cross corresponding to the peak you just added
Confirm the correct row has been removed from the table
The cross should be removed from the plot
Repeat the above steps 1-7 in non-orthogonal view.
Check the
Concise Viewcheckbox above one of the peak tables on the right hand side - this should hide some columns.
MDHistoWorkspace¶
Make a 3D MDHistoWorkspace
md_3D_histo = BinMD(InputWorkspace='md_4D', AlignedDim0='H,-2,2,100', AlignedDim1='K,-1,1,100', AlignedDim2='L,-1.5,1.5,100')
Open
md_3D_histoin sliceviewer it should not support dynamic rebinning (can’t change number of bins).Test the toolbar buttons pan, zoom, line plots, ROI as in step 3 of the MatrixWorkspace instructions.
Test changing/swapping viewing axes
Test the Nonorthogonal view as above
Open
md_4D_svrebinnedin sliceviewer (should be in the ADS after preceding tests).It should support dynamic rebinning (i.e. will be able to change number of bins along each axis).
With
md_4D_svrebinnedopen in the sliceviewer, deletemd_4Din the ADS.It should close sliceviewer because the support for dynamic rebinning has changed
Open
md_4D_svrebinnedin sliceviewer againIt should no longer support dynamic rebinning
Confirm transposing axes works
CutViewer Tool¶
1. Check the Non-axis aligned cutting tool button is only enabled for 3D MD workspaces where all dimensions are Q by opening the following workspaces in sliceviewer. It should only be enabled for the ws_3D and ws_3D_QLab workspaces (see comment for details) - the first 3 column headers for the vectors should be a*,b*,c* and Qx,Qy,Qz for the two workspaces respectively.
Load(Filename='CNCS_7860_event.nxs', OutputWorkspace='CNCS_7860_event') # disabled (MatrixWorkspace)
ws_2D = CreateMDWorkspace(Dimensions='2', Extents='-5,5,-4,4', Names='H,K',
Units='r.l.u.,r.l.u.', Frames='HKL,HKL',
SplitInto='2', SplitThreshold='50') # disabled (2D MD)
ws_3D = CreateMDWorkspace(Dimensions='3', Extents='-5,5,-4,4,-3,3',
Names='H,K,L', Units='r.l.u.,r.l.u.,r.l.u.',
Frames='HKL,HKL,HKL', SplitInto='2', SplitThreshold='50') # enabled!
ws_3D_nonQdim = CreateMDWorkspace(Dimensions=3, Extents=[-1, 1, -1, 1, -1, 1],
Names="E,H,K", Frames='General Frame,HKL,HKL',
Units='meV,r.l.u.,r.l.u.') # disabled (3D but 1 non-Q)
ws_4D = CreateMDWorkspace(Dimensions=4, Extents=[-1, 1, -1, 1, -1, 1, -1, 1],
Names="E,H,K,L", Frames='General Frame,HKL,HKL,HKL',
Units='meV,r.l.u.,r.l.u.,r.l.u.') # disabled (4D - one non-Q)
ws_3D_QLab = CreateMDWorkspace(Dimensions='3', Extents='-5,5,-4,4,-3,3',
Names='Q_lab_x,Q_lab_y,Q_lab_z', Units='U,U,U',
Frames='QLab,QLab,QLab', SplitInto='2', SplitThreshold='50') # enabled!
Close any sliceviewer windows and clear the workspaces in the ADS
Run the following and open ws in sliceviewer.
ws = CreateMDWorkspace(Dimensions='3', Extents='-5,5,-4,4,-3,3',
Names='H,K,L', Units='r.l.u.,r.l.u.,r.l.u.',
Frames='HKL,HKL,HKL', SplitInto='2', SplitThreshold='50')
expt_info = CreateSampleWorkspace()
SetUB(expt_info, 1,1,2,90,90,120)
ws.addExperimentInfo(expt_info)
# make some fake data
FakeMDEventData(ws, UniformParams='1e5', PeakParams='1e+05,0,0,1,0.3', RandomSeed='3873875')
Open the cut tool and check that:
Disabled ROI tool
Disabled line plots
Sliceviewer should look like
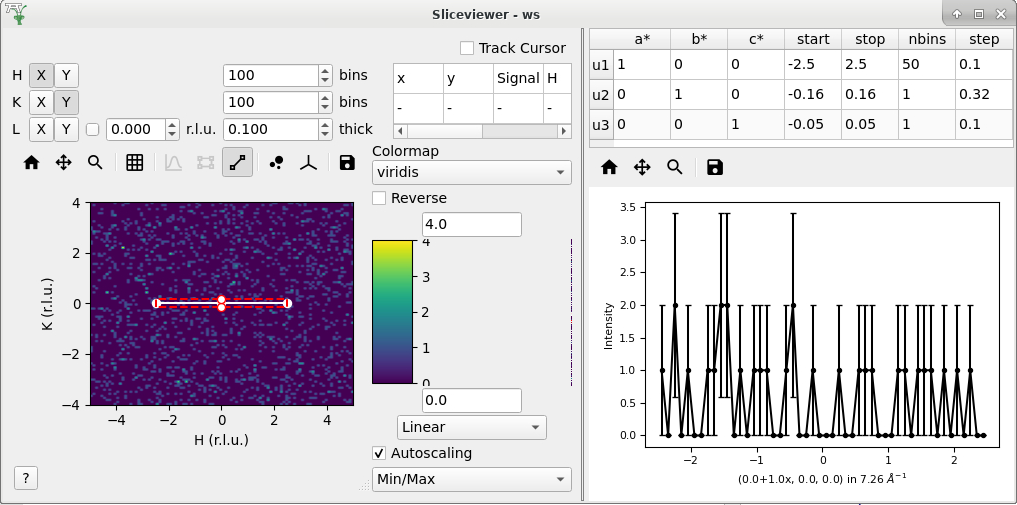
Check that transposing the axes (X <-> Y) will swap the u1 and u2 vectors in the table
Set the axes to (X,Y) = (L, K) - check u1 = [0,0,1] and u2 = [0,1,0]
Change the slice point of H to be 0 - it should look like this
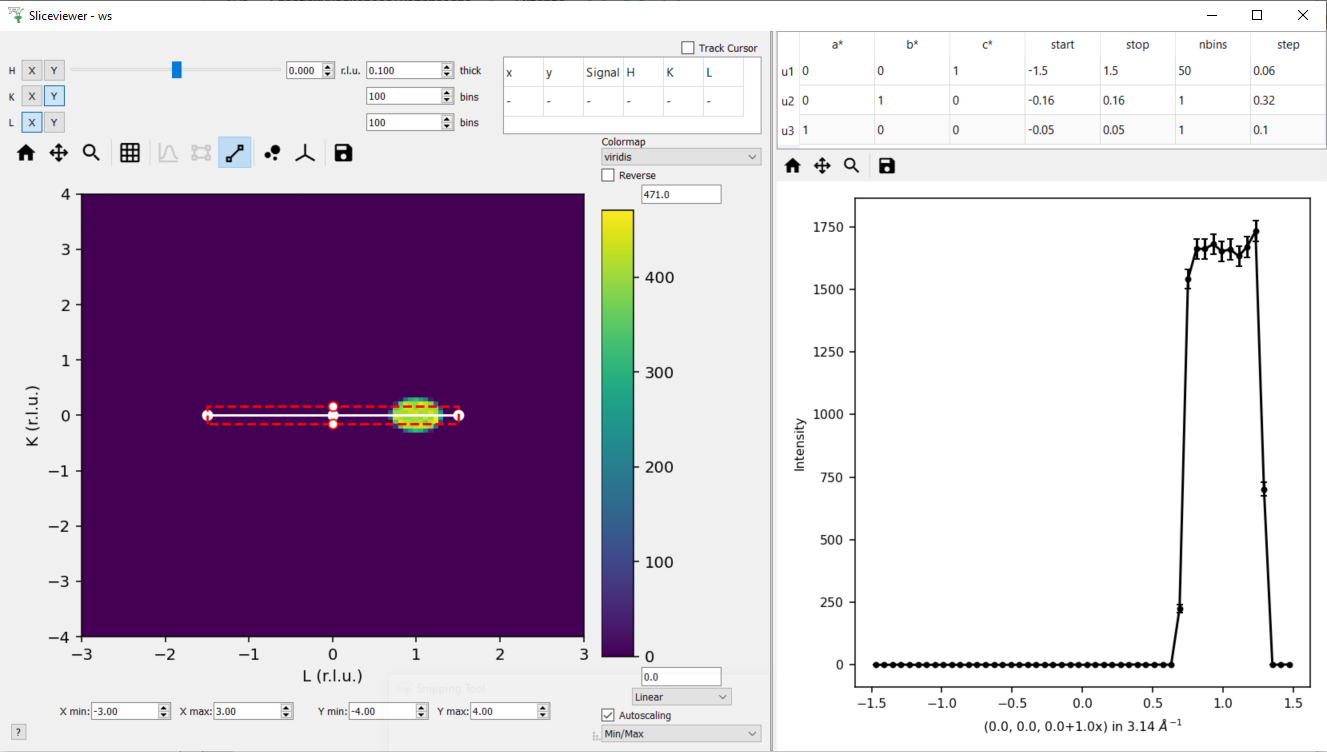
In the table double the step along u1 (i.e. set step to 0.12) - this should change nbins = 25 along u1
Set the nbins = 50 along u1 - the step should go back to it’s original value (0.06)
Set the stop for u1 to 0 and check that
step size = 0.03
the cut representation line on the colorfill plot has the correct start/stop
the 1D plot in the cut viewer pane has the correct axes limits
Set step of u1 to be 2 (i.e. greater than the extent of the cut) - this should set nbins=1 and step = 1.5 and put the cut along u2 with nbins = 50.
Transpose the axes so now (X,Y) = (K,L) - the cut should have 50 bins along K (default value)
Change the nbins of u2 to 50 (it should set nbins=1 for u1 and change the step=4). Check the white line of the cut representation on the colorfill plot is now vertical.
Try to change the a* column of the u1 to 1 (this would take u1 out of the plane of the slice, i.e. not orthogonal to u3) - it should reset to 0 - i.e. u1 = [0,1,0].
Click and hold down on one of the red markers with white face on the colorfill plot and drag, release at ~K=1.
This should reset the vectors in the table such that the cut is along u1 = [0,0,1] - i.e. u1 and u2 are swapped
The thickness along u2 should be adjusted to ~2
Set the step of u2 = 2 in the table, check that it sets (start,stop) = (-1,1)
Drag the top white marker of the cut representation up to L~2
u1 ~ [0,0,1] and u2 ~ [0,1,0]
There should be a peak in the 1D plot at x~1
For u1 set b*=0 and c* = -1 and start=-2 - check that u2 = [0,-1,0] and there is a peak at x~-1
To change the centre of the cut move the central white marker of the cut representation to (K,L) ~ (2,0),
The entire cut representation should move
The axes label of the 1D plot should be similar to \((0.0, 2.0,0.0-1.0x) \ in \ 3.14 \AA^{-1}\)
There should be no peak on the 1D plot
Increase the thickness by dragging the left red marker of the cut representation to encompass the peak in the data - check the peak appears in the 1D plot at the right thickness.
Play around with the direction of the cut by dragging the white markers at the end points of the white line - the vectors u1 and u3 should be orthogonal unit vectors.
Reset the cut by transposing the axes so (X,Y) = (L,K)
Double the slice thickness along H from 0.1 -> 0.2 (the counts of the peak in the 1D plot should double from ~1750 -> ~3500)
Change c* of u1 from 1 -> -1
The peak in the 1D plot should move from x= 1 -> -1
Check u2 = [0,-1,0]
Turn on non-orthogonal view and play around with the cut tool representation
Check that the table values are updated correctly (use the HKL in the cursor info table to help determine this)
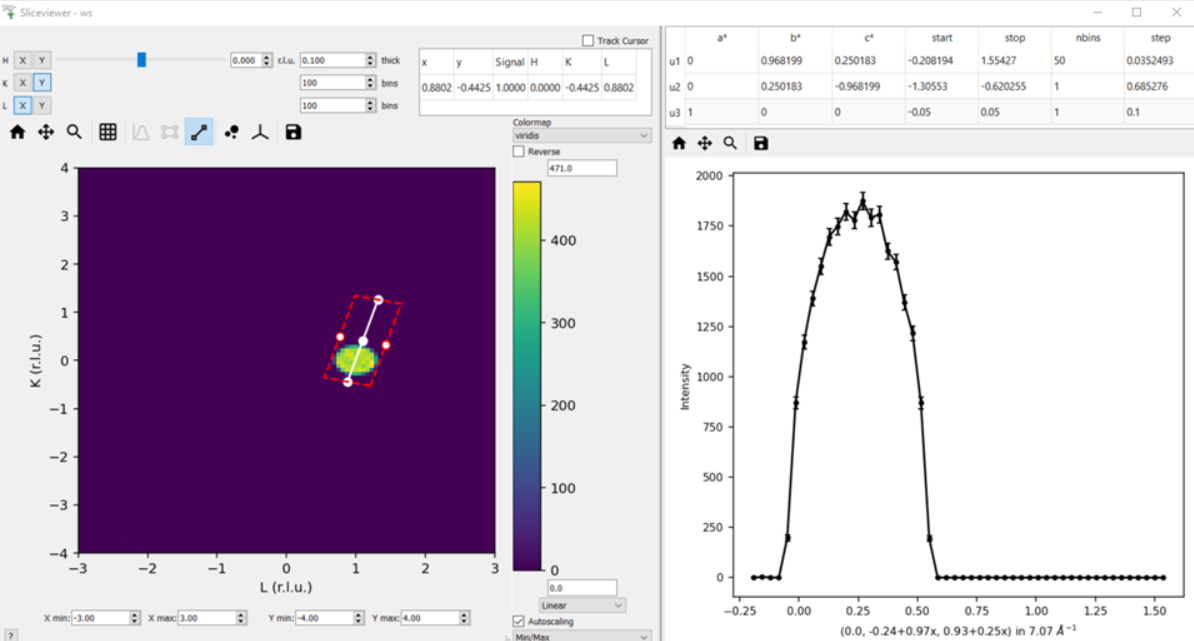
Produce a non-axis aligned cut as shown below where the peak is not in the center of the x-range. Take the x-value at the peak maximum and plug it into the axis label - it should produce the HKL of the peak (0,0,1).
In the example below, the x axis label is \((0.0, -0.24 + 0.97x, 0.93 + 0.25x) \ in \ 7.07 \AA^{-1}\). So if we plug in the value of x at the peak centre we get
(0.0, -0.24 + 0.97*0.25, 0.93 + 0.25*0.25)=(0.0, 0.0025, 0.9925), which is very close to actual peak HKL =(0,0,1)
Specific Tests¶
1. Representation of integrated peaks¶
Run the code below to generate fake data and integrate peaks in the 3D MDWorkspace
md_3Dfrom MDWorkspace (with events)
# Fake data in 3D MD and integrate
FakeMDEventData(md_3D, EllipsoidParams='1e4,1,0,1,1,0,0,0,1,0,0,0,1,0.005,0.005,0.015,0', RandomSeed='3873875') # ellipsoid
FakeMDEventData(md_3D, EllipsoidParams='1e4,1,0,0,1,0,0,0,1,0,0,0,1,0.005,0.005,0.005,0', RandomSeed='3873875') # spherical
IntegratePeaksMD(InputWorkspace='md_3D', PeakRadius='0.25', BackgroundInnerRadius='0.25', BackgroundOuterRadius='0.32', PeaksWorkspace='peaks', OutputWorkspace='peaks_int_ellip', IntegrateIfOnEdge=False, Ellipsoid=True, UseOnePercentBackgroundCorrection=False)
IntegratePeaksMD(InputWorkspace='md_3D', PeakRadius='0.25', BackgroundInnerRadius='0.25', BackgroundOuterRadius='0.32', PeaksWorkspace='peaks', OutputWorkspace='peaks_int_sphere', IntegrateIfOnEdge=False, Ellipsoid=False, UseOnePercentBackgroundCorrection=False)
IntegratePeaksMD(InputWorkspace='md_3D', PeakRadius='0.25', BackgroundInnerRadius='0', BackgroundOuterRadius='0', PeaksWorkspace='peaks', OutputWorkspace='peaks_int_no_bg', IntegrateIfOnEdge=False, Ellipsoid=False, UseOnePercentBackgroundCorrection=False)
# IntegratePeaksMD will throw an error
# Error in execution of algorithm MaskBTP:...
# This is because the simulated ws don't have a real instrument but the integration will be executed
Open
md_3Din sliceviewerClick the peak overlay button in the toolbar
Overlay
peaks_int_ellipandpeaks_int_sphereClick the first row in the first table
It should zoom to a peak.
There should be an ellipse and a circle drawn with dashed lines with different colors (the color should match the color of the workspace name in the peak viewer table).
There should be a transparent shell indicating the background for each peak.
The ellipse should be smaller than the circle.
Alter the slice point by moving the slider along the integrated dimension
The circle and ellipse should shrink
There should be no gap between the background shell and the dashed line.
Click on the second row on the second table.
It should zoom in on a different peak.
The ellipse and circle should be very similar (not quite same as the covariance matrix was evaluated numerically for randomly generated data).
Click the nonorthogonal view button
The ellipse and circle should still agree with each other and the shape of the generated data.
Click the Peak overlay button in the toolbar
Overlay the
peaks_int_no_bgworkspace and removepeaks_int_sphereZoom in on a peak (click a row in the table)
There should be a dashed line but no background shell for peaks in
peaks_int_no_bg
Keep the three peak workspaces overlain for the next test.
2. ADS observer for peak overlay¶
Rename
peaks_int_ellipin the ADS to e.g.peaks_int_ellipseConfirm the name changes in the peak viewer table
Click on a peak, the ellipse should still be drawn on the colofill plot
Remove a row from
peaks_int_no_bgtable (open table from ADS > Right-click on a row > Delete)Confirm the correct row is removed from the corresponding row in the peak viewer table
Click on the peak in the
peaks_int_ellipsetable that has been removed frompeaks_int_no_bgOnly the ellipse should be plotted.
Delete
peaks_int_no_bgfrom the ADSThe table should be removed from the peaks viewer
Confirm the Peak actions combo box is updated to only contain
peaks_int_ellipse
Delete
peaks_int_ellipsefrom the ADSThe peak overlay should be turned off and the table hidden
3. ADS observer for workspace¶
With md_3D open in sliceviewer
Rename
md_3Dto e.g.md_3DimThe workspace name in the title of the sliceviewer window should have updated
Zoom to check dynamic rebinning still works
Ensure colorbar autoscale is checked.
Take a note of the colorbar limits and execute this command in the ipython terminal
mtd['md_3Dim'] *= 2
The colorbar max should be doubled.
Zoom to check dynamic rebinning still works
Clone the workspace for future tests
CloneWorkspace(InputWorkspace='md_3Dim', OutputWorkspace='md_3D')
Delete
md_3Dimin the ADSThe sliceviewer window should close
4. ADS observer for support for nonorthogonal view¶
Open
md_3Din sliceviewerRun
ClearUBalgorithm onmd_3DSliceviewer window should close with message
Closing Sliceviewer as the underlying workspace was changed: The property supports_nonorthogonal_axes is different on the new workspace.
5. Check BinMD called with NormalizeBasisVectors=False for HKL data¶
Create a workspace with peaks at integer HKL and take a non axis-aligned cut
ws = CreateMDWorkspace(Dimensions='3', Extents='-3,3,-3,3,-3,3',
Names='H,K,L', Units='r.l.u.,r.l.u.,r.l.u.',
Frames='HKL,HKL,HKL',
SplitInto='2', SplitThreshold='10')
# add fake Bragg peaks for primitive lattice in data
for h in range(-3,4):
for k in range(-3,4):
for l in range(-3,4):
hkl = ",".join([str(x) for x in [h,k,l]])
FakeMDEventData(ws, PeakParams='1e+02,' + hkl + ',0.02', RandomSeed='3873875')
BinMD(InputWorkspace=ws, AxisAligned=False,
BasisVector0='[00L],U,0,0,1',
BasisVector1='[HH0],U,1,1,0',
BasisVector2='[-HH0],U,-1,1,0',
OutputExtents='-4,4,-4,4,-0.25,0.25',
OutputBins='101,101,1', OutputWorkspace='BinMD_out', NormalizeBasisVectors=False)
Open
BinMD_outin sliceviewer.There should be peaks at integer HKL
Zoom in (so that the data are rebinned)
The peaks should still be at integer HKL (rather than multiples of \(\sqrt{2}\))
6. Check gets the correct basis vectors for MDHisto workspaces¶
This tests that the sliceviewer gets the correct basis vectors for an MDHisto object from a non-axis aligned cut.
Create the workspace
ws = CreateMDWorkspace(Dimensions='3', Extents='-3,3,-3,3,-3,3',
Names='H,K,L', Units='r.l.u.,r.l.u.,r.l.u.',
Frames='HKL,HKL,HKL',
SplitInto='2', SplitThreshold='10')
expt_info = CreateSampleWorkspace()
ws.addExperimentInfo(expt_info)
SetUB(ws, 1,1,2,90,90,120)
BinMD(InputWorkspace=ws, AxisAligned=False,
BasisVector0='[00L],r.l.u.,0,0,1',
BasisVector1='[HH0],r.l.u.,1,1,0',
BasisVector2='[-HH0],r.l.u.,-1,1,0',
OutputExtents='-4,4,-4,4,-0.25,0.25',
OutputBins='101,101,1', OutputWorkspace='ws_slice', NormalizeBasisVectors=False)
Open
ws_slicein the sliceviewer.The non-orthogonal view should be enabled (not greyed out).
Click the non-orthogonal view button
Rectangular gridlines should appear (as in this case 110 is orthogonal to 001).
7. Check non-orthogonal view is disabled for non-Q axes¶
Check that the non-orthogonal view is disabled for non-Q axes such as energy
Create a workspace with energy as the first axis.
ws_4D = CreateMDWorkspace(Dimensions=4, Extents=[-1, 1, -1, 1, -1, 1, -1, 1], Names="E,H,K,L",
Frames='General Frame,HKL,HKL,HKL', Units='meV,r.l.u.,r.l.u.,r.l.u.')
expt_info_4D = CreateSampleWorkspace()
ws_4D.addExperimentInfo(expt_info_4D)
SetUB(ws_4D, 1, 1, 2, 90, 90, 120)
Open
ws_4Din sliceviewer.Confirm that when the Energy axis is viewed (as X or Y) the non-orthogonal view is disabled.
The button should be re-enabled when you view two Q-axes e.g. H and K.